Elemental sulfur (S0) and reduced inorganic sulfur compounds (RISCs) such as hydrogen sulfide or thiosulfate are abundant resources for energy conservation of microbes throughout the biosphere. Sulfur compounds are found in almost all habitats however, they are especially important in freshwater lakes, oceans and their sediments, in hydrothermal volcanic areas and biotechnologically, in the extraction of metals from ores, mining slag heaps and waste streams (bioleaching).
Model organisms and their enzymes
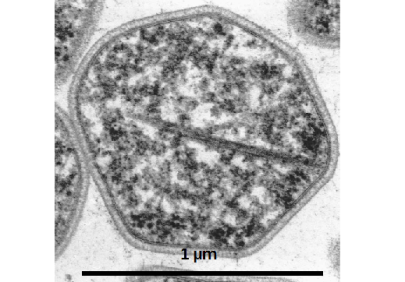
Acidianus ambivalens, the “oldest” and best-studied of our model Archaea, grows in volcanic hot springs and fumaroles at 80˚C and pH 2.5 utilizing S0 and other sulfur compounds as sole energy sources either aerobically or anaerobically with H2 as electron donor. During aerobic growth, S0 is oxidized to sulfuric acid which causes the acidity to drop below pH 1. During anaerobic growth, copious amounts of H2S are formed. In addition, Acd. ambivalens uses sulfuric metal ores such as pyrite (FeS2), chalcopyrite (CuFeS2) and other minerals as sole energy sources.
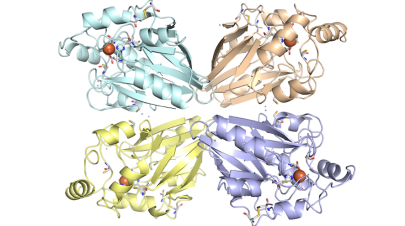
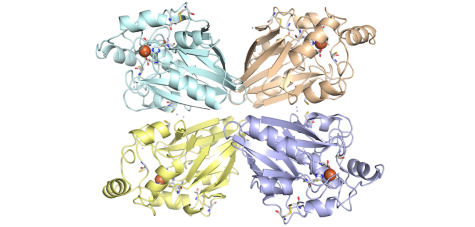
We had purified and characterized several enzymes from this Archaeon, especially sulfur oxygenase reductase (SOR), for which we solved the three-dimensional structure, but also enzymes such as hydrogenase and sulfur reductase, thiosulfate:quinone oxidoreductase, and tetrathionate hydrolase.
Sulfur reduction and cytochromes c in Ignicoccus hospitalis:
The distantly related and strictly anaerobic archaeon I. hospitalis (Topt = 90˚C, pHopt = 5.5) also grows by H2 oxidation coupled to S0 reduction. It was initially used as another model organism for sulfur reduction, however, we switched to studying the abundant cytochromes c. We could show that three different multiheme cytochromes c can be purified out of four predicted from the genome sequence of the organisms. Unfortunately, it remained unclear how they participated in sulfur metabolism. During these studies we conducted a survey on cytochromes c and found them being present in many – but far from all – methanogens, and anaerobic sulfate, sulfur and iron-reducing Archaea suggesting different biochemical pathways depending on the respective species.
Persulfide dioxygenase from Acidithiobacillus caldus:
Act. caldus is a moderately thermophilic and acidophilic bacterium widely found in bioleaching environments. It also has a sor gene but we were interested in another enzyme, initially described as sulfur dioxygenase, which later turned out to be a persulfide dioxygenase instead (PDO). It uses glutathione persulfide to oxidize the sulfur to sulfite.
Thioalkalivibrio paradoxus sulfur oxygenase and proteome studies:
A different “lifestyle” is followed by the natrono-alkaliphilic bacterium Thioalkalivibrio paradoxus (opens in new tab) isolated from soda lakes – it grows aerobically around 30-37˚C at pH 10 oxidizing not only sulfide, S0 and thiosulfate but also thiocyanate. It also has sor- and pdo- genes. When purifying the SOR, it turned out that the enzyme has different properties compared to the A. ambivalens enzyme.
Gold bioleaching and cyanide-producing bacteria
Mobile phones and other electronic devices contain gold, cobalt and other metals, little of which is currently recovered. In spite of its toxicity, cyanide is still the most common chemical for gold dissolution from mining ores and electronic waste. Other studies had shown, that cyanide-producing bacteria are efficient in gold recovery from waste, while producing and utilizing much less of the toxic compound compared to chemical extraction. It is right no problematic that the model organisms used so far for cyanide bioleaching, Chromobacterium violaceum and Pseudomonas aeruginosa, are potentially pathogenic even apart from cyanide production.
In order to find non-pathogenic cyanide producers that can be applied industrially, we currently isolate many cyanide producing bacteria and determine their biotechnological potential. In addition, we try to isolate the cyanide-producing enzyme, which is biochemically not known at all.
Techniques
- Bacterial isolation and characterization
- Heterologous gene expression, mutagenesis of the respective genes
- Biochemical characterization of proteins, especially metal-containing proteins
- 3D structure prediction
- 3D structure determination in collaboration with Carlos Frazão (ITQB, Oeiras, Portugal)
- Proteome studies of Thioalkalivibrio sp. in collaboration with Christof Lenz (MPI Biophysical Chemistry, Göttingen, Ger.)