Respiratory reduction of laughing gas
An environmentally important molecule of the biogeochemical nitrogen cycle is nitrous oxide (N2O, laughing gas), which is a potent greenhouse gas and an ozone depleting substance. We study the N2O metabolism of nitrate/nitrite-ammonifying and denitrifying bacteria that are capable to grow by nitrous oxide respiration. A prime example for this type of energy metabolism is the non-pathogenic rumen bacterium Wolinella succinogenes. As this bacterium is known to generate small amounts of nitric oxide (NO) and nitrous oxide during nitrate ammonification, we also explore the enzymology of nitric oxide detoxification and nitrosative stress defence.
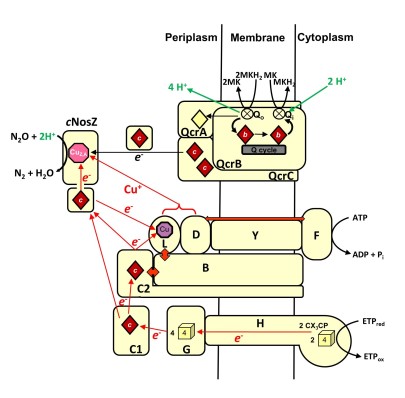
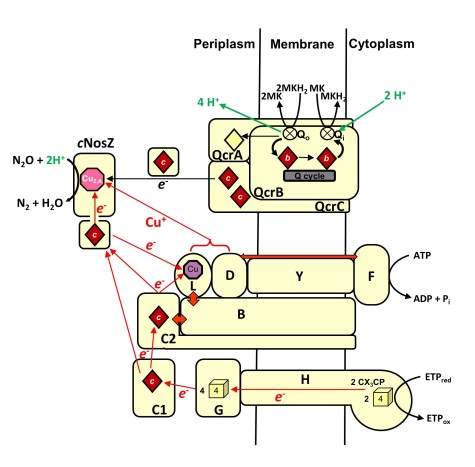
The terminal reductase of W. succinogenes nitrous oxide respiration is a cytochrome c nitrous oxide reductase (cNosZ) that belongs to the so-called clade II of NosZ enzymes. We presented evidence that functional N2O respiration involves a menaquinol-oxidizing Rieske/cytochrome bc complex and various Nos proteins encoded in the nos gene cluster. Furthermore, transcriptome analysis suggested that N2O-grown W. succinogenes cells specifically regulate gene expression in response to nitrous oxide.
Over the last century the atmospheric concentration of nitrous oxide has been constantly on the rise. This fact is caused by the invention of man-made nitrogen fixation (the Haber-Bosch process), which led to a drastic intensification of agricultural fertilization practices and a severe anthropogenic imbalance of the global biogeochemical nitrogen cycle. In a biotechnological approach we aim to apply suitable N2O-respiring bacteria to different natural habitats to reduce N2O emissions.
We use a wide range of methods in cellular and molecular microbiology in combination with genetic engineering of suitable model bacteria, omics techniques, protein biochemistry and structural biology.
MccA-dependent sulphite respiration
Sulphite is reduced to sulfide by the highly active octahaem cytochrome c sulphite reductase MccA, present in the periplasm of certain Proteobacteria. The high-resolution crystal structure of Wolinella succinogenes MccA revealed an unprecedented haem c-copper active site of sulfite reduction and confirmed the presence of a haem c group bound to a unique CX15CH haem attachment site.
Currently, we explore the constituents of the corresponding electron transport chain and the process of MccA maturation, which is supposed to include the putative copper chaperone MccL.
Biosynthesis and function of methylated menaquinone derivatives
The membranous quinone/quinol pool is an essential component of most bacterial and archaeal respiratory electron transport chains. Beside the well-known ubiquinone and menaquinone (MK), many bacteria produce mono- and dimethylated MK derivatives such as 8-methyl-MK and 7,8-dimethyl-MK.
We identified and characterized a family of HemN-related class C radical S-adenosylmethionine methyltransferases designated MenK, MenK2 or MqnK that methylate MK specifically at position C-8 (MenK/MqnK) or C-7 (MenK2). The MenK and MenK2 enzymes from Adlercreutzia equolifaciens were functionally produced in Wolinella succinogenes or Escherichia coli cells. We currently explore the reaction mechanism of these enzymes and the cellular function of methylated menaquinones.
Open positions
We are seeking highly motivated students at the BSc, MSc and PhD level. Projects for Bachelor and Master theses for Biology, Biochemistry and Biomolecular Engineering students are offered in the research areas described above. Please contact Jörg Simon for details on current projects.